Overview
STellaris is a platform designed for rapid spatial mapping for single-cell RNA sequencing (scRNA-seq) data based on emerging spatial transcriptomics (ST) data. Our goal is to help users make full use of the increasing volume of single-cell omics data in the spatial context. We provide walk-through tutorials on the three major tools implemented in STellaris, including spatial mapping, dataset browserand gene search tools.
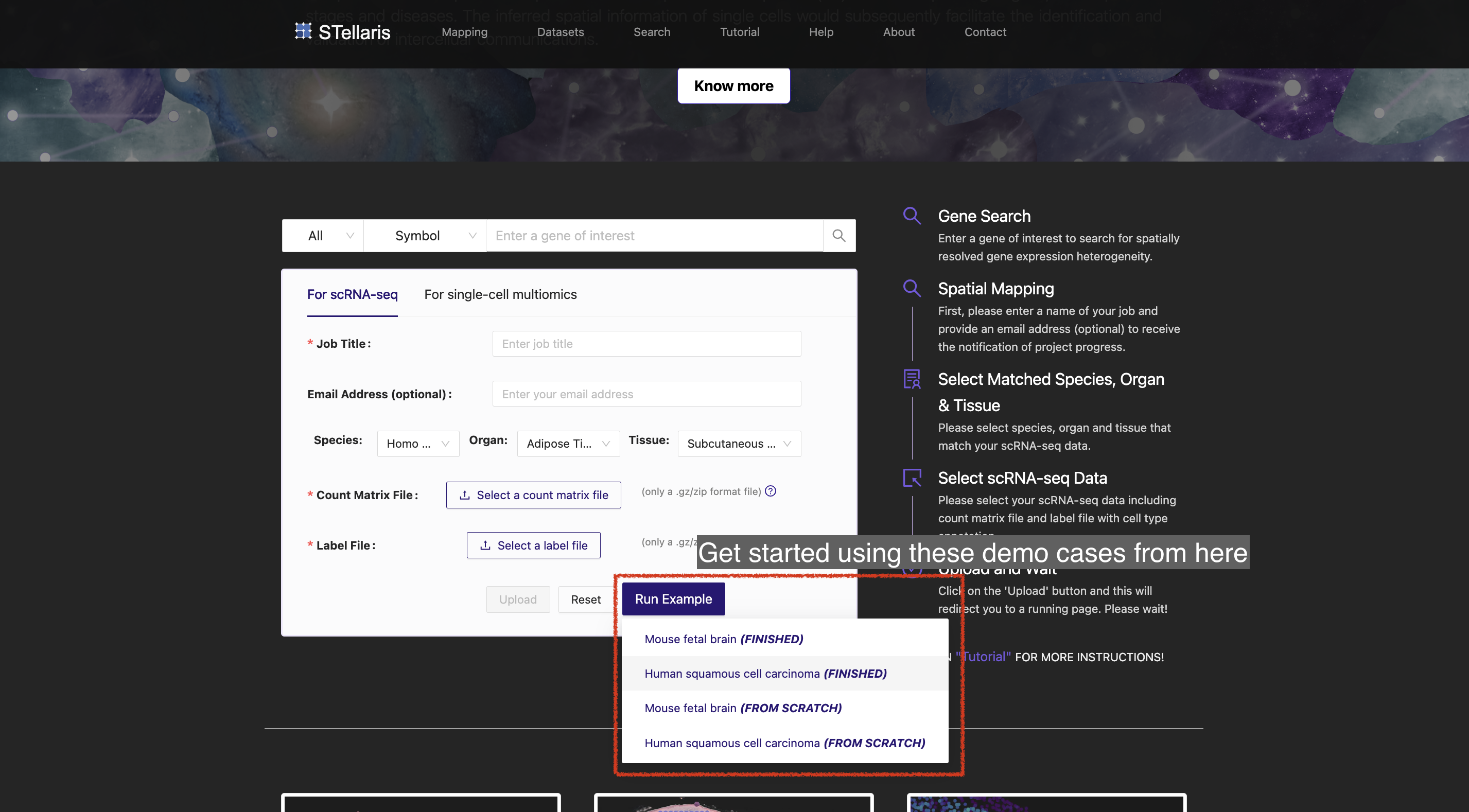
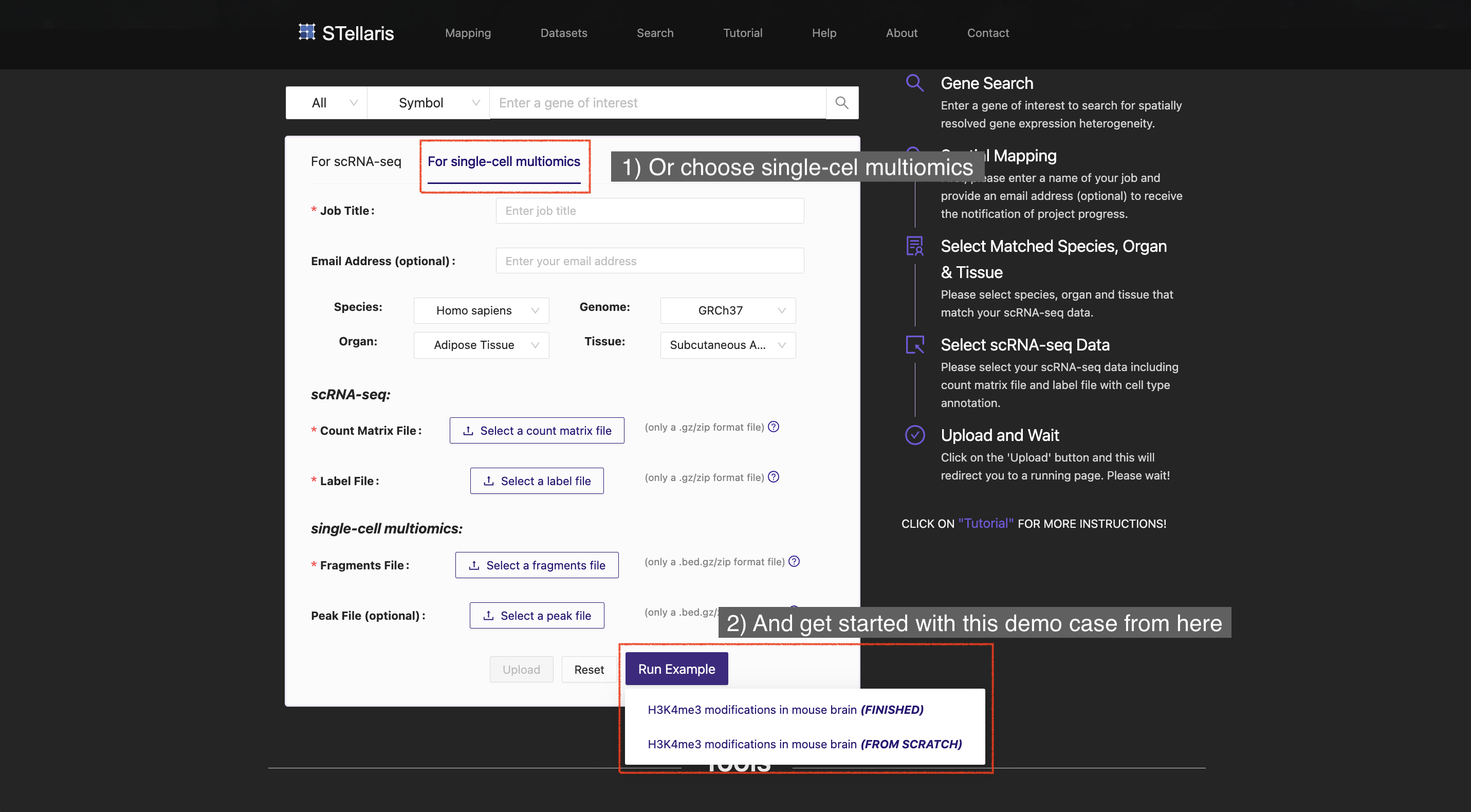
We offer guidance to users on how to get started to map their annotated scRNA-seq data to spatial positions in tissue sections curated in our local database (see Get started). We also provide a tutorial on how to systematically interpret the spatial mapping results and downstream intercellular communication results. (see Result interpretation). To demonstrate the usefulness of STellaris in real-world data, we provide tutorials on the case studies: (i) dissecting the spatial organization of the developing mouse cerebral cortex (see Mouse fetal brain), (ii) mapping intercellular crosstalk at tumor leading edges of human squamous cell carcinoma (see Human squamous cell carcinoma), and (iii) deciphering the spatial patterning of H3K4me3 modification in the mouse brain (see H3K4me3 modification in mouse brain). In addition, we offer tutorials on how to view our curated ST datasets (Dataset browser) and retrieve tissue-wide expression signatures of genes (Gene search) from a spatial perspective.
Please note that we provide three case studies that have already been completed:
Additionally, we offer three corresponding job request that can be executed to repeat these results:
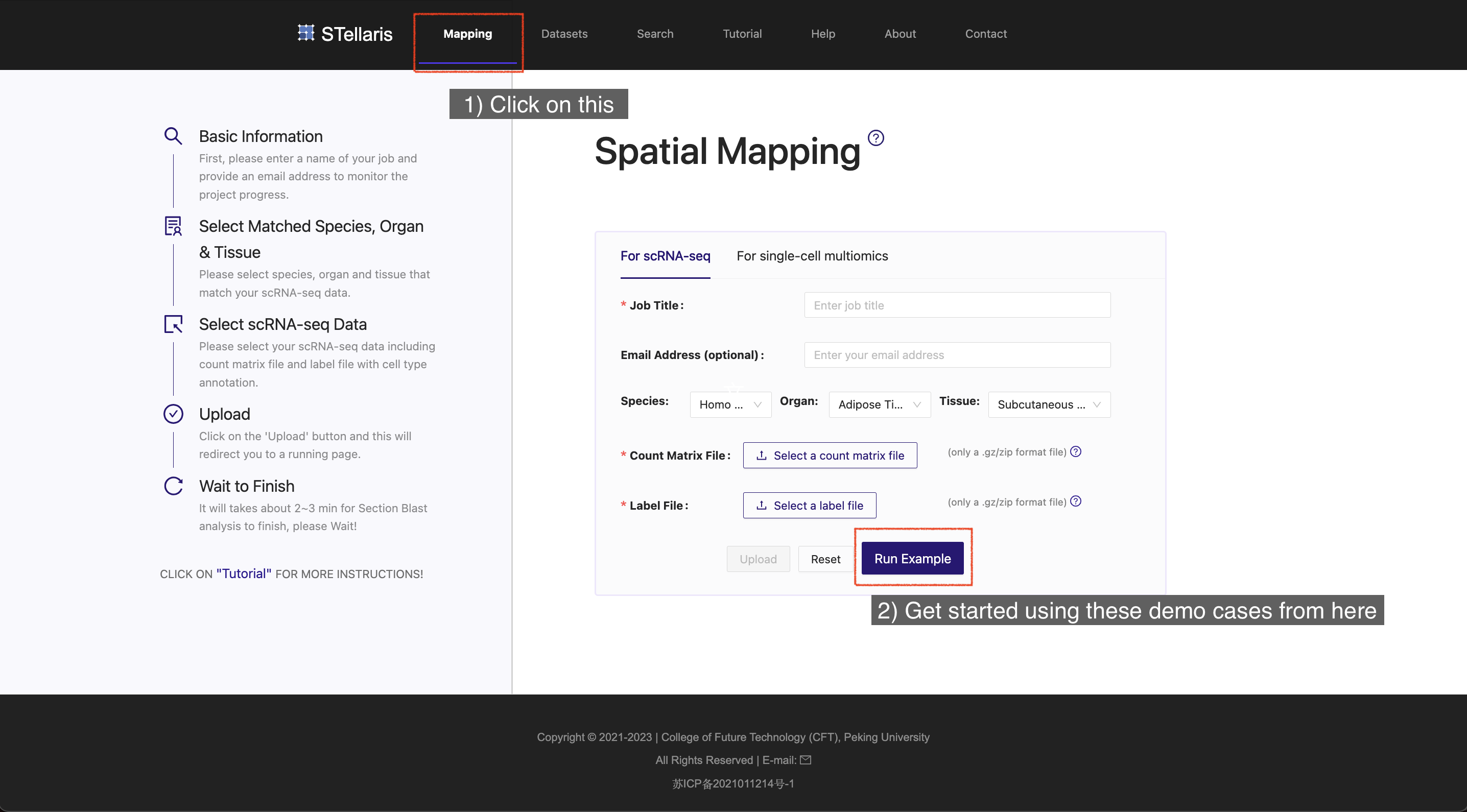
To explore these demo cases, simply visit the Home page or Mapping page in the navigation bar.